
Anatomy of a Data Product — Part Five
This is an article in a series about building data products with Tag.bio.
To begin the series, check out Part One, which outlines the reason for and definition of a data product, along with key concepts and terms. To access the data & codebase to follow along with these examples, see Part Two.
In Part Four we introduced protocols — registered, searchable API methods which invoke customizable queries, algorithms and visualizations on the data model (established by the config in Part Three).
Here, in Part Five, we will show how protocol functionality can be extended with R & Python plugins and present an example R plugin protocol from fc-iris-demo.

The example R plugin protocol is located in the fc-iris-demo project at protocols/protocol_r_plotly.json.
R plugin protocols are mostly the same as native ones
If you recall Part Four, in the section describing protocol_definition, you may note that the protocol_definition of the R plugin protocol above contains the same attributes as a native protocol:
- name
- title
- description
- asset
- argument_sets
R & Python plugin protocols are designed to be used and invoked in exactly the same way as native protocols — i.e. the User Experience is the same. Here’s what the protocol configuration screen looks like from the UI:

What’s special about an R plugin protocol?
There are two distinct differences between an R protocol and a native protocol:
- script — the method attribute tells the system to invoke R (or Python).
- protocol_output — there is no protocol_output attribute for an R protocol, as the output from R (or Python) is directly returned as the API response.
Script
Let’s drill into the script section of the example R plugin protocol:

Here, the script section contains six attributes:
- method — the value is “external”, which tells the system to invoke R (or Python).
- sdk — this tells the system which external environment to use.
- plugin — a file path to the R plugin code for this protocol.
- output_type — the value is “html”, which prepares the API response to return an HTML output. Alternative options here include: “png”, “svg”, and “pdf”.
- background — as described in Part Four, this will prepare a data frame for the plugin using this set of entities as rows.
- analysis_variables — as described in Part Four, this will specify the columns of the data frame for the plugin with collections of variables.
The plugin function

Each R (or Python) plugin contains a single function that will be executed when the protocol is invoked from the API. The function is required to have an explicit parameter signature, accepting two arguments:
- tag_data — contains the data frame prepared by the protocol, an authentication token for the user invoking the protocol, and argument values specified in the API request.
- tag_result — an object for storing the output from the plugin. In this case, the output is a plotly (HTML+JS+CSS) file.
Upon invocation, the plugin function uses the data and parameters provided in tag_data to execute algorithms and produce one or more visualizations to be stored in tag_output.
The plugin function output is then returned as the API response:

The R Markdown variant
The fc-iris-demo example data product contains another R plugin protocol which operates in a slightly different way. The protocol JSON is essentially the same, but the R plugin code is different.
This example is located at protocols/protocol_r_markdown.json.

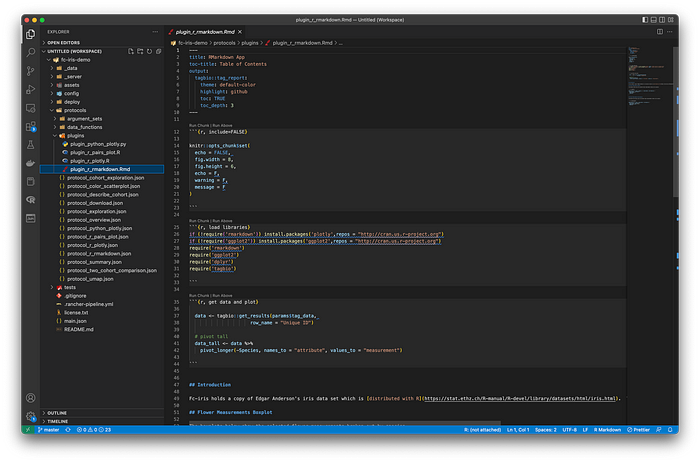
Note how the plugin file extension (.Rmd), and the structure of the code is different from the previously-described plugin. This is a pure R Markdown script — but it still has access to tag_data in the execution environment to receive the data frame, user auth token, and argument values from the protocol. No tag_result environment variable is required, because the code is automatically rendered into HTML+JS+CSS using R Markdown.
The output of an R Markdown protocol, using the Tag.bio rendering theme, looks like this:


It’s a powerful way to create data products with customizable, detailed analysis reports generated from precise algorithms and visualizations.
Acknowledgements
Firstly — thank you, reader — for taking the time to read and learn about data products within our framework. Please reach out in the comments or send an email to info@tag.bio if you have any questions or feedback. Or visit the Tag.bio website to learn more about research and business applications.
Data product developers:
Wade Webster, Daniel Warren, J Ireland, Susann Edler-Childress, Fleur Leenen, Rocio Dominguez Vidana, Tom Paquette
Front end designers & developers:
Ames Cornish, Jan Simmala, Georgi Serev, Katerina Skroumpelou
Platform management & cloud engineers:
Sanjay Padhi, Derek De Jonghe, Kenn Brodhagen
Product leadership:
Tom Covington, Mark Mooney
