
Anatomy of a Data Product — Part Two
This is an article in a series about building data products with Tag.bio.
To begin the series, check out Part One, which outlines the reason for and definition of a data product, along with key concepts and terms.
Here, in Part Two, we will introduce the example dataset and provide a link to the corresponding data product codebase so you can follow along with the code in the rest of the series.
Fisher’s Iris Dataset
We’ve chosen an extremely simple and well-known data source, so we can focus on the technology behind the data product, instead of the data itself.
You will not need to download the data on your own—it will be provided when you clone the git repository (explained below).

Fischer’s Iris Dataset contains petal and sepal dimension measurements for 150 individual flowers. The data source is a single table, encoded as a CSV file, with 6 columns:
- observation — a unique ID for each flower
- petal_length
- petal_width
- sepal_length
- sepal_width
- species — each flower belongs to one of 3 different iris species, with 50 flowers belonging to each category
Here’s what the dataset looks like as a table:

The fc-iris-demo Data Product
As discussed in Part One, a Data Product is comprised of two digital components:
- A data snapshot
- A codebase
The codebase contains low-code instructions — a config — telling the Tag.bio framework how to connect to the source dataset(s), and how to ingest and model the data into a snapshot. The snapshot is then stored on a file system.
The codebase also contains low-code protocols and plugins which define the API methods that the data product will provide when deployed.
Here’s how to access the codebase for this project, fc-iris-demo. Side note — “fc” stands for “Flux Capacitor”, the legacy name for our data product framework.
- You can view the code in your browser from: https://bitbucket.org/protocolbuilders/fc-iris-demo/src/master/
- Or — better yet — you can clone the Git repository by running this command from a terminal:
git clone https://bitbucket.org/protocolbuilders/fc-iris-demo.git
Working with a Data Product in a coding environment (IDE)
If you clone the git repository to your local machine, you will want to view the contents from a code editor (IDE). At Tag.bio, we tend to use either Visual Studio Code, or RStudio. Both are (generally) free to download and use.
Note: screenshots and code examples in the rest of this series will be provided from Visual Studio Code.
One important tip for using Visual Studio Code — our .json files contain comments, so you will need to configure the IDE to use a jsonc editor for .json files. To confirm or change your setting, open the main.json file in the fc-iris-demo project and check this line at the bottom of the screen.

It should show “JSON with Comments”, not “JSON”. To change this from JSON, click on JSON in the bottom bar and choose Configure File Association for ‘.json’… from the pop up menu.
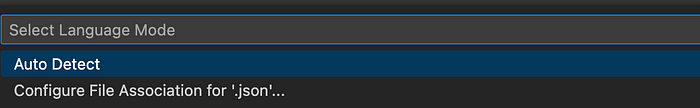
Then, choose JSON with Comments from the pop-up menu.

The main.json file should look like the screenshot below (if you’re using dark mode, that is), and show no syntax errors (red lines at the far right of the screen).

Take note that the _data/ folder in the fc-iris-demo project holds the file data.csv, which contains Fisher’s Iris Dataset. Data Products typically never include source data files inside the git repository, but for the simplicity of this tutorial we’re making an exception.

What’s next?
Part Three in this series will show how the low-code config defines data ingestion, modeling, and snapshot creation from Fisher’s Iris Dataset.
